Take a Deep Dive into Your Scientific Imaging Data
arivis Vision4D 3.5 already proved that you do not need to be an expert in bioinformatics to unleash the power of machine learning (ML) for advanced results in less time. The newest version of our leading software platform for scientific image visualization and analysis goes even further. Vision4D 3.6 for the first time features a seamless and flexible workflow for deep learning (DL) applications with multilayered neural networks for faster-than-ever results from your valuable data. Well-trained deep learning algorithms can save you precious time and effort when annotating and segmenting even the most demanding samples from your instrument such as 3D FIB-SEM and CryoEM volume imaging data. Deep Learning can speed up the time required for tedious analysis tasks from several weeks to mere hours, freeing up valuable resources in your research lab for more important tasks.
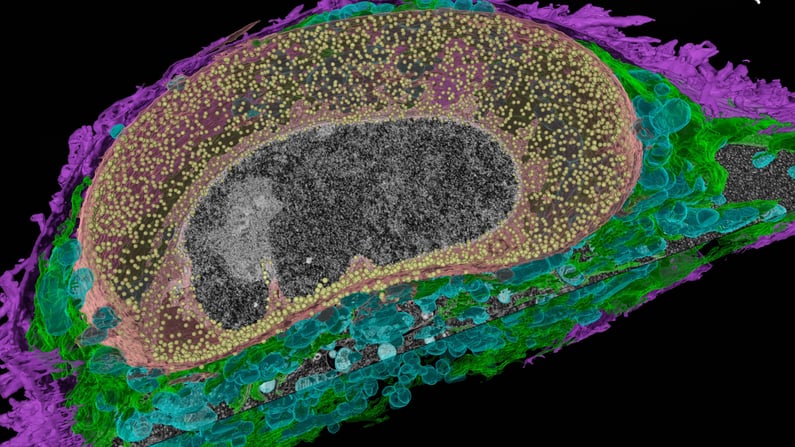 Automated 3D serial imaging of HeLa cells with ZEISS Crossbeam FIB-SEM. The focused ion beam was utilized to sequentially remove 8 nm thick layers of the specimen while the exposed block-face is scanned with a scanning electron microscope, thus obtaining a high-resolution 3D volume image. Automated segmentation and visualization of cellular components with an APEER-trained deep learning model in Vision4D 3.6. Dataset kindly provided by Anna Steyer and Yannick Schwab, EMBL Heidelberg, Germany.
Automated 3D serial imaging of HeLa cells with ZEISS Crossbeam FIB-SEM. The focused ion beam was utilized to sequentially remove 8 nm thick layers of the specimen while the exposed block-face is scanned with a scanning electron microscope, thus obtaining a high-resolution 3D volume image. Automated segmentation and visualization of cellular components with an APEER-trained deep learning model in Vision4D 3.6. Dataset kindly provided by Anna Steyer and Yannick Schwab, EMBL Heidelberg, Germany.
Top Features of arivis Vision4D 3.6:
Vision4D 3.6 fully supports import of deep learning models in the Open Neural Network Exchange (ONNX) format as well as the CZ model from ZEISS ZEN Intellesis and the APEER cloud-based image processing platform. With APEER you easily annotate and train new models or combine already existing ones into customized workflows, without the need for programming knowledge*. Vision4D now gives you the flexibility to import and apply fully trained neural networks from various libraries and backends to your analysis pipelines to enable deep learning for automated analysis of your imaging data.

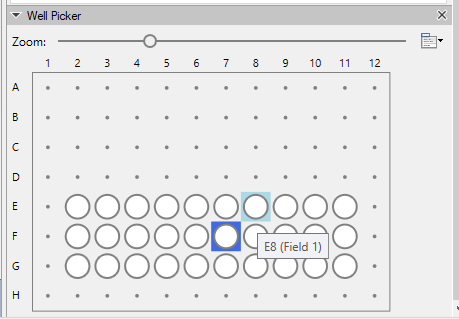
Additionally, Vision4D now features improved support for well plate imaging datasets with a new graphical overview, easy navigation and intuitive selection of the position of your well plates image set. Advanced visualization and automated analysis of your high-content imaging applications has never been more efficient! Fully supported formats and systems include ZEISS Celldiscoverer 7 and any CZI file with well plate information as well as selected Molecular Devices and Yokogawa imaging systems**. Further updates to Vision4D 3.6 include the Blob Finder analysis tool that now comes with more options and parameters for normalization as well as various bug fixes and speed improvements across the board.
We make sure that with arivis Vision4D you always stay at the forefront of multidimensional image visualization and analysis with the most advanced technologies, workflows and tools. The arivis scientific imaging platform offers a seamless integration with VisionHub for storage and processing of vast amounts of imaging data and VisionVR, the unique virtual reality experience for immersive visualization, analysis, education and collaboration.
arivis Vision4D 3.6 yet again boosts your scientific imaging research: Visualize, process and get your results as efficiently and comfortable as never before! Scroll down to contact us for a remote demonstration or get your free trial license today!
*) Free for academic users. Price models for commercial users available at www.apeer.com/biotech
**) For a detailed list of supported vendors and formats get in contact with our specialists.
Existing V4D users with a "Maintenance Contract" license can directly download and install the latest update ("Help" --> "Check for update").
Please make sure that your workstation is connected to the Internet and your Firewall is not blocking the application. If in doubt please contact your administrator or your arivis Imaging Specialist. Detailed technical release notes can be found in our arivis Knowledge Base here.
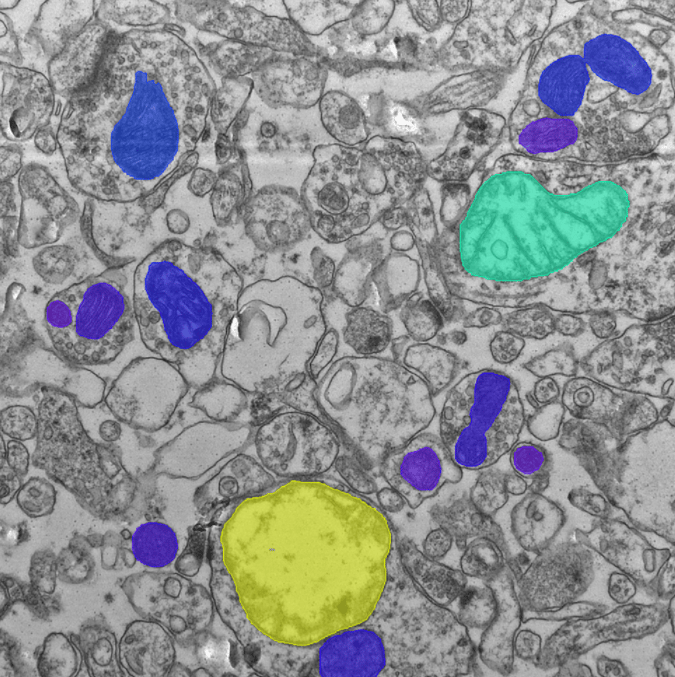
Hippocampus tissue section, transmission electron microscopy. In total, 30 TEM serial sections were used with 309 mitochondria objects, annotated manually. Mitochondria were segmented with an ONNX Deep Learning model that was imported and applied in Vision4D 3.6 (YouTube video tutorial). Original imaging data kindly provided by Dr. Wendy Bautista, MD PhD, Barrow Neurological Institute, Phoenix Children’s Hospital.
