arivis Scientific Image Analysis
Close
- Products
- Applications
- Resources
- Publications
- Case Studies
- Mitochondria Analysis with Deep Learning
- Compartmentalization of Chromatin
- Gastrulation & Migration Phenotypes
- Advanced 4D Tracking of Meiotic Spindle Dynamic
- Migration of Interneurons in Developing Mouse Brain
- Exploring Whole Organisms at Single-cell Resolution
- Image Analysis Workflow for EM images
- Webinars and Workshops
- Tutorials
- AI eBook
- AI Blog
- How to's
- Service & Support
- About
- Products
- Applications
- Resources
- Publications
- Case Studies
- Mitochondria Analysis with Deep Learning
- Compartmentalization of Chromatin
- Gastrulation & Migration Phenotypes
- Advanced 4D Tracking of Meiotic Spindle Dynamic
- Migration of Interneurons in Developing Mouse Brain
- Exploring Whole Organisms at Single-cell Resolution
- Image Analysis Workflow for EM images
- Webinars and Workshops
- Tutorials
- AI eBook
- AI Blog
- How to's
- Service & Support
- About


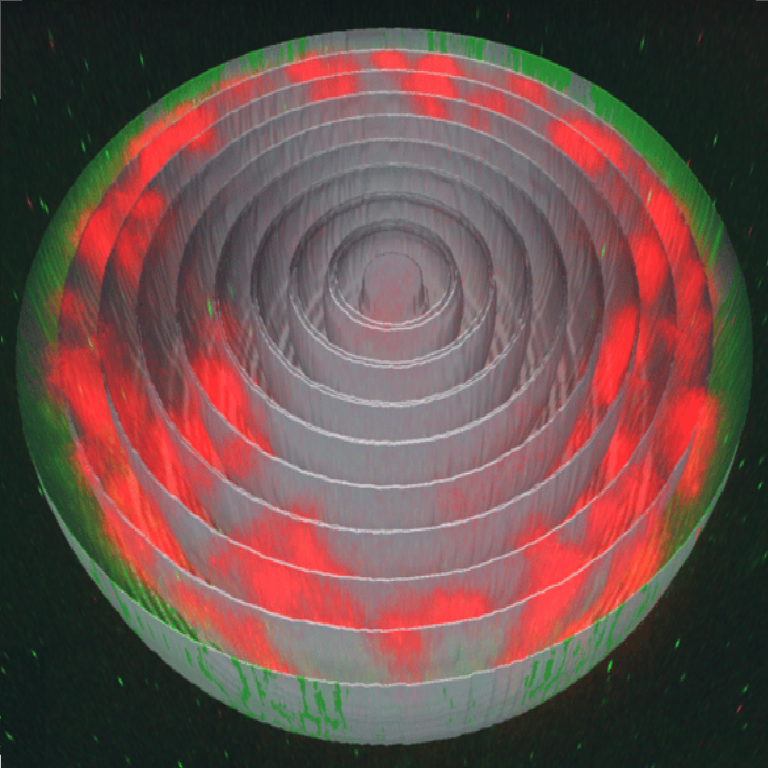


-1.png?width=1920&name=CONTROL-180719_muscle_klar_H2B_nuc3-highres%20(2)-1.png)